Segment the objects from the plane, color them¶
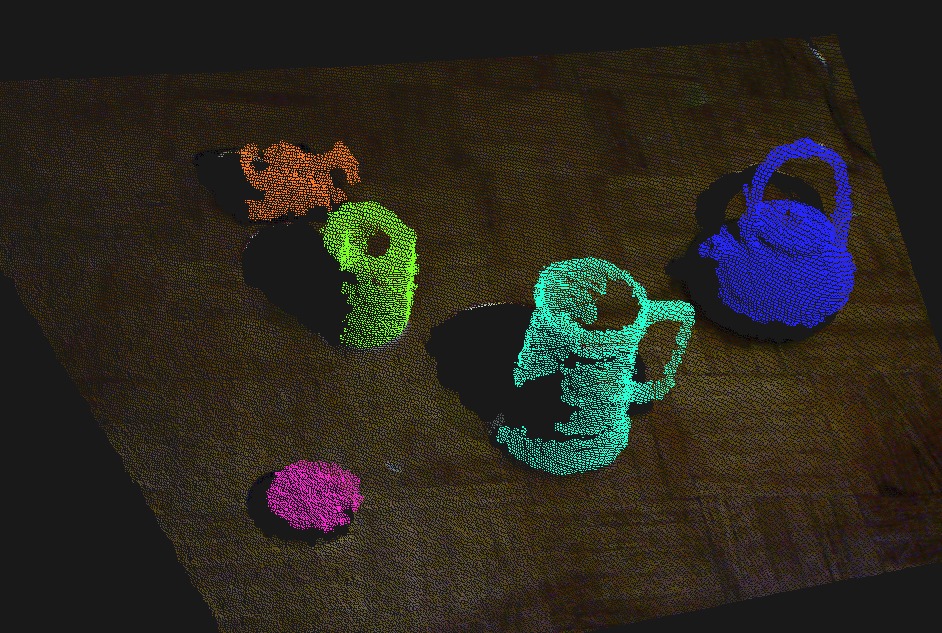
#!/usr/bin/env python
"""
This example shows how to extract points corresponding to objects on a table,
cluster them, colorize the clusters, and republish as a single cloud.
1) The example downsamples using a VoxelGrid before estimating
normals for the downsampled cloud.
2) These normals are then used for segmentation using RANSAC.
3) Segmentation produces a planar model to which all inliers are
projected so that a 2D convex hull can be created.
4) We then extract the indices of all points that are above the
plane formed by the convex hull.
5) This cloud is then clustered
6) The clusters are concatenated, with each having a unique color.
"""
import ecto
from ecto.opts import run_plasm, scheduler_options
from ecto_openni import Capture
from ecto_pcl import *
# capture from kinect and downsample
device = Capture('device')
cloud_generator = NiConverter('cloud_generator')
voxel_grid = VoxelGrid("voxel_grid", leaf_size=0.05)
graph = [device[:] >> cloud_generator[:],
cloud_generator[:] >> voxel_grid[:]
]
# estimate normals, segment and find convex hull
normals = NormalEstimation("normals", k_search=0, radius_search=0.2)
planar_segmentation = SACSegmentationFromNormals("planar_segmentation",
model_type=SACMODEL_NORMAL_PLANE,
eps_angle=0.09, distance_threshold=0.1)
project_inliers = ProjectInliers("project_inliers", model_type=SACMODEL_NORMAL_PLANE)
nan_filter = PassThrough('nan_removal')
convex_hull = ConvexHull("convex_hull")
graph += [voxel_grid[:] >> normals[:],
voxel_grid[:] >> planar_segmentation["input"],
normals[:] >> planar_segmentation["normals"],
voxel_grid[:] >> project_inliers["input"],
planar_segmentation["model"] >> project_inliers["model"],
project_inliers[:] >> nan_filter[:],
nan_filter[:] >> convex_hull['input']
]
# extract stuff on table from original high-res cloud, find clusters, colorize, merge and show in viewer
extract_stuff = ExtractPolygonalPrismData("extract_stuff", height_min=0.01, height_max=0.2)
extract_indices = ExtractIndices("extract_indices", negative=False)
extract_clusters = EuclideanClusterExtraction("extract_clusters", min_cluster_size=50, cluster_tolerance=0.005)
colorize = ColorizeClusters("colorize")
merge = MergeClouds("merge")
viewer = CloudViewer("viewer", window_name="Clouds!")
graph += [cloud_generator[:] >> extract_stuff["input"],
convex_hull[:] >> extract_stuff["planar_hull"],
extract_stuff[:] >> extract_indices["indices"],
cloud_generator[:] >> extract_indices["input"],
extract_indices[:] >> extract_clusters['input'],
extract_clusters[:] >> colorize["clusters"],
extract_indices[:] >> colorize["input"],
cloud_generator[:] >> merge["input"],
colorize[:] >> merge["input2"],
colorize[:] >> viewer[:]
]
plasm = ecto.Plasm()
plasm.connect(graph)
if __name__ == "__main__":
from ecto.opts import doit
doit(plasm, description='Execute tabletop segmentation and colorize clusters.')
![digraph G {
graph [rankdir=TB, ranksep=1]
edge [labelfontsize=8]
node [shape=plaintext]
0[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR> <TD ROWSPAN="9" COLSPAN="7" BGCOLOR="khaki">device</TD> <TD PORT="p_depth_fps" BGCOLOR="lightblue">depth_fps</TD>
</TR> <TR> <TD PORT="p_depth_resolution" BGCOLOR="lightblue">depth_resolution</TD> </TR>
<TR> <TD PORT="p_device" BGCOLOR="lightblue">device</TD> </TR>
<TR> <TD PORT="p_device_number" BGCOLOR="lightblue">device_number</TD> </TR>
<TR> <TD PORT="p_device_uid" BGCOLOR="lightblue">device_uid</TD> </TR>
<TR> <TD PORT="p_registration" BGCOLOR="lightblue">registration</TD> </TR>
<TR> <TD PORT="p_rgb_fps" BGCOLOR="lightblue">rgb_fps</TD> </TR>
<TR> <TD PORT="p_rgb_resolution" BGCOLOR="lightblue">rgb_resolution</TD> </TR>
<TR> <TD PORT="p_synchronize" BGCOLOR="lightblue">synchronize</TD> </TR>
<TR>
<TD PORT="o_depth_buffer" BGCOLOR="indianred1">depth_buffer</TD>
<TD PORT="o_depth_height" BGCOLOR="indianred1">depth_height</TD>
<TD PORT="o_depth_width" BGCOLOR="indianred1">depth_width</TD>
<TD PORT="o_image_buffer" BGCOLOR="indianred1">image_buffer</TD>
<TD PORT="o_image_channels" BGCOLOR="indianred1">image_channels</TD>
<TD PORT="o_image_height" BGCOLOR="indianred1">image_height</TD>
<TD PORT="o_image_width" BGCOLOR="indianred1">image_width</TD>
</TR> </TABLE>>];
1[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_depth_buffer" BGCOLOR="springgreen">depth_buffer</TD>
<TD PORT="i_depth_height" BGCOLOR="springgreen">depth_height</TD>
<TD PORT="i_depth_width" BGCOLOR="springgreen">depth_width</TD>
<TD PORT="i_image_buffer" BGCOLOR="springgreen">image_buffer</TD>
<TD PORT="i_image_channels" BGCOLOR="springgreen">image_channels</TD>
<TD PORT="i_image_height" BGCOLOR="springgreen">image_height</TD>
<TD PORT="i_image_width" BGCOLOR="springgreen">image_width</TD>
</TR> <TR> <TD ROWSPAN="1" COLSPAN="7" BGCOLOR="khaki">cloud_generator</TD> </TR> <TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
2[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
</TR> <TR> <TD ROWSPAN="5" COLSPAN="1" BGCOLOR="khaki">voxel_grid</TD> <TD PORT="p_filter_field_name" BGCOLOR="lightblue">filter_field_name</TD>
</TR> <TR> <TD PORT="p_filter_limit_max" BGCOLOR="lightblue">filter_limit_max</TD> </TR>
<TR> <TD PORT="p_filter_limit_min" BGCOLOR="lightblue">filter_limit_min</TD> </TR>
<TR> <TD PORT="p_filter_limit_negative" BGCOLOR="lightblue">filter_limit_negative</TD> </TR>
<TR> <TD PORT="p_leaf_size" BGCOLOR="lightblue">leaf_size</TD> </TR>
<TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
3[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
</TR> <TR> <TD ROWSPAN="6" COLSPAN="1" BGCOLOR="khaki">normals</TD> <TD PORT="p_k_search" BGCOLOR="lightblue">k_search</TD>
</TR> <TR> <TD PORT="p_radius_search" BGCOLOR="lightblue">radius_search</TD> </TR>
<TR> <TD PORT="p_spatial_locator" BGCOLOR="lightblue">spatial_locator</TD> </TR>
<TR> <TD PORT="p_vp_x" BGCOLOR="lightblue">vp_x</TD> </TR>
<TR> <TD PORT="p_vp_y" BGCOLOR="lightblue">vp_y</TD> </TR>
<TR> <TD PORT="p_vp_z" BGCOLOR="lightblue">vp_z</TD> </TR>
<TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
4[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_indices" BGCOLOR="springgreen">indices</TD>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
<TD PORT="i_normals" BGCOLOR="springgreen">normals</TD>
</TR> <TR> <TD ROWSPAN="13" COLSPAN="3" BGCOLOR="khaki">planar_segmentation</TD> <TD PORT="p_axis_x" BGCOLOR="lightblue">axis_x</TD>
</TR> <TR> <TD PORT="p_axis_y" BGCOLOR="lightblue">axis_y</TD> </TR>
<TR> <TD PORT="p_axis_z" BGCOLOR="lightblue">axis_z</TD> </TR>
<TR> <TD PORT="p_distance_threshold" BGCOLOR="lightblue">distance_threshold</TD> </TR>
<TR> <TD PORT="p_eps_angle" BGCOLOR="lightblue">eps_angle</TD> </TR>
<TR> <TD PORT="p_max_iterations" BGCOLOR="lightblue">max_iterations</TD> </TR>
<TR> <TD PORT="p_method" BGCOLOR="lightblue">method</TD> </TR>
<TR> <TD PORT="p_model_type" BGCOLOR="lightblue">model_type</TD> </TR>
<TR> <TD PORT="p_normal_distance_weight" BGCOLOR="lightblue">normal_distance_weight</TD> </TR>
<TR> <TD PORT="p_optimize_coefficients" BGCOLOR="lightblue">optimize_coefficients</TD> </TR>
<TR> <TD PORT="p_probability" BGCOLOR="lightblue">probability</TD> </TR>
<TR> <TD PORT="p_radius_max" BGCOLOR="lightblue">radius_max</TD> </TR>
<TR> <TD PORT="p_radius_min" BGCOLOR="lightblue">radius_min</TD> </TR>
<TR>
<TD PORT="o_inliers" BGCOLOR="indianred1">inliers</TD>
<TD PORT="o_model" BGCOLOR="indianred1">model</TD>
</TR> </TABLE>>];
5[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
<TD PORT="i_model" BGCOLOR="springgreen">model</TD>
</TR> <TR> <TD ROWSPAN="2" COLSPAN="2" BGCOLOR="khaki">project_inliers</TD> <TD PORT="p_copy_all_data" BGCOLOR="lightblue">copy_all_data</TD>
</TR> <TR> <TD PORT="p_model_type" BGCOLOR="lightblue">model_type</TD> </TR>
<TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
6[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
</TR> <TR> <TD ROWSPAN="4" COLSPAN="1" BGCOLOR="khaki">nan_removal</TD> <TD PORT="p_filter_field_name" BGCOLOR="lightblue">filter_field_name</TD>
</TR> <TR> <TD PORT="p_filter_limit_max" BGCOLOR="lightblue">filter_limit_max</TD> </TR>
<TR> <TD PORT="p_filter_limit_min" BGCOLOR="lightblue">filter_limit_min</TD> </TR>
<TR> <TD PORT="p_filter_limit_negative" BGCOLOR="lightblue">filter_limit_negative</TD> </TR>
<TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
7[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_indices" BGCOLOR="springgreen">indices</TD>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
</TR> <TR> <TD ROWSPAN="1" COLSPAN="2" BGCOLOR="khaki">convex_hull</TD> <TD PORT="p_dimensionality" BGCOLOR="lightblue">dimensionality</TD>
</TR> <TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
8[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
<TD PORT="i_planar_hull" BGCOLOR="springgreen">planar_hull</TD>
</TR> <TR> <TD ROWSPAN="2" COLSPAN="2" BGCOLOR="khaki">extract_stuff</TD> <TD PORT="p_height_max" BGCOLOR="lightblue">height_max</TD>
</TR> <TR> <TD PORT="p_height_min" BGCOLOR="lightblue">height_min</TD> </TR>
<TR>
<TD PORT="o_inliers" BGCOLOR="indianred1">inliers</TD>
</TR> </TABLE>>];
9[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_indices" BGCOLOR="springgreen">indices</TD>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
</TR> <TR> <TD ROWSPAN="2" COLSPAN="2" BGCOLOR="khaki">extract_indices</TD> <TD PORT="p_keep_organized" BGCOLOR="lightblue">keep_organized</TD>
</TR> <TR> <TD PORT="p_negative" BGCOLOR="lightblue">negative</TD> </TR>
<TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
10[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_indices" BGCOLOR="springgreen">indices</TD>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
</TR> <TR> <TD ROWSPAN="3" COLSPAN="2" BGCOLOR="khaki">extract_clusters</TD> <TD PORT="p_cluster_tolerance" BGCOLOR="lightblue">cluster_tolerance</TD>
</TR> <TR> <TD PORT="p_max_cluster_size" BGCOLOR="lightblue">max_cluster_size</TD> </TR>
<TR> <TD PORT="p_min_cluster_size" BGCOLOR="lightblue">min_cluster_size</TD> </TR>
<TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
11[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_clusters" BGCOLOR="springgreen">clusters</TD>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
</TR> <TR> <TD ROWSPAN="3" COLSPAN="2" BGCOLOR="khaki">colorize</TD> <TD PORT="p_max_clusters" BGCOLOR="lightblue">max_clusters</TD>
</TR> <TR> <TD PORT="p_saturation" BGCOLOR="lightblue">saturation</TD> </TR>
<TR> <TD PORT="p_value" BGCOLOR="lightblue">value</TD> </TR>
<TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
12[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
<TD PORT="i_input2" BGCOLOR="springgreen">input2</TD>
</TR> <TR> <TD ROWSPAN="1" COLSPAN="2" BGCOLOR="khaki">merge</TD> </TR> <TR>
<TD PORT="o_output" BGCOLOR="indianred1">output</TD>
</TR> </TABLE>>];
13[label=<<TABLE BORDER="0" CELLBORDER="1" CELLSPACING="0" CELLPADDING="4"> <TR>
<TD PORT="i_input" BGCOLOR="springgreen">input</TD>
</TR> <TR> <TD ROWSPAN="1" COLSPAN="1" BGCOLOR="khaki">viewer</TD> <TD PORT="p_window_name" BGCOLOR="lightblue">window_name</TD>
</TR> </TABLE>>];
0->1 [headport="i_depth_buffer" tailport="o_depth_buffer"];
0->1 [headport="i_depth_height" tailport="o_depth_height"];
0->1 [headport="i_depth_width" tailport="o_depth_width"];
0->1 [headport="i_image_buffer" tailport="o_image_buffer"];
0->1 [headport="i_image_channels" tailport="o_image_channels"];
0->1 [headport="i_image_height" tailport="o_image_height"];
0->1 [headport="i_image_width" tailport="o_image_width"];
1->2 [headport="i_input" tailport="o_output"];
2->3 [headport="i_input" tailport="o_output"];
2->4 [headport="i_input" tailport="o_output"];
3->4 [headport="i_normals" tailport="o_output"];
2->5 [headport="i_input" tailport="o_output"];
4->5 [headport="i_model" tailport="o_model"];
5->6 [headport="i_input" tailport="o_output"];
6->7 [headport="i_input" tailport="o_output"];
1->8 [headport="i_input" tailport="o_output"];
7->8 [headport="i_planar_hull" tailport="o_output"];
8->9 [headport="i_indices" tailport="o_inliers"];
1->9 [headport="i_input" tailport="o_output"];
9->10 [headport="i_input" tailport="o_output"];
10->11 [headport="i_clusters" tailport="o_output"];
9->11 [headport="i_input" tailport="o_output"];
1->12 [headport="i_input" tailport="o_output"];
11->12 [headport="i_input2" tailport="o_output"];
11->13 [headport="i_input" tailport="o_output"];
}](../_images/graphviz-4208ce26e32e365bd7ddce5f183912b61b3dddc0.png)